KiPhyNet
An online network simulation tool connecting cellular kinetics and physiological transport.
KiPhyNet is a web application that facilitates the simulation and visualization
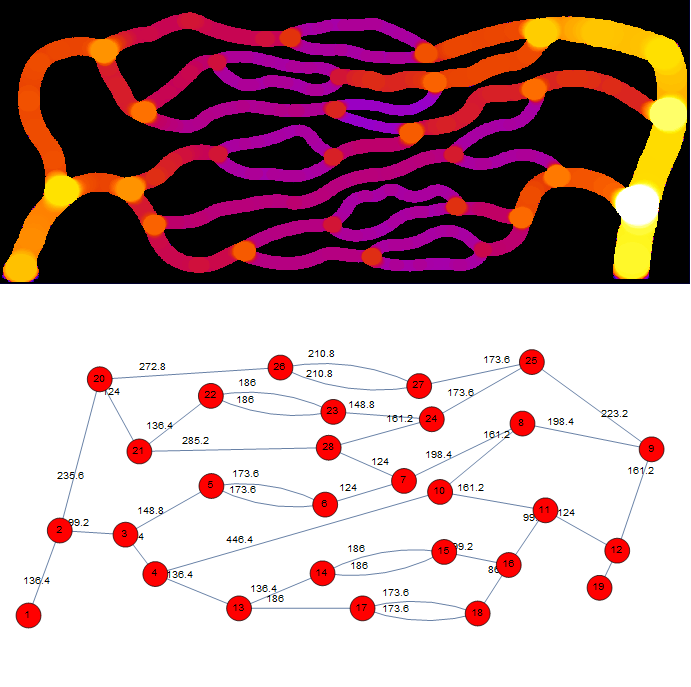
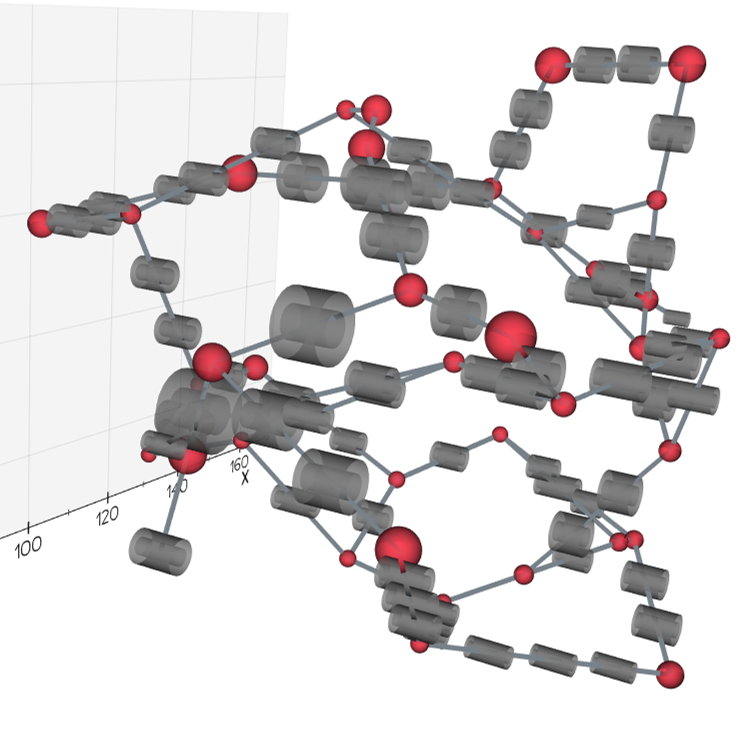
of advection-dispersion transport in three-dimensional (3D) microvascular networks. KiPhyNet provides the functionality to
assimilate multi-omics data from clinical and experimental studies as well as vascular data from imaging studies to
investigate the role of structural changes in vascular topology on the functional response of the tissue.
KiPhyNet is implemented in Python based on Apache web server using MATLAB as the simulator engine.
With the network topology, its biophysical attributes, and the values of the kinetic and transport constants, a user
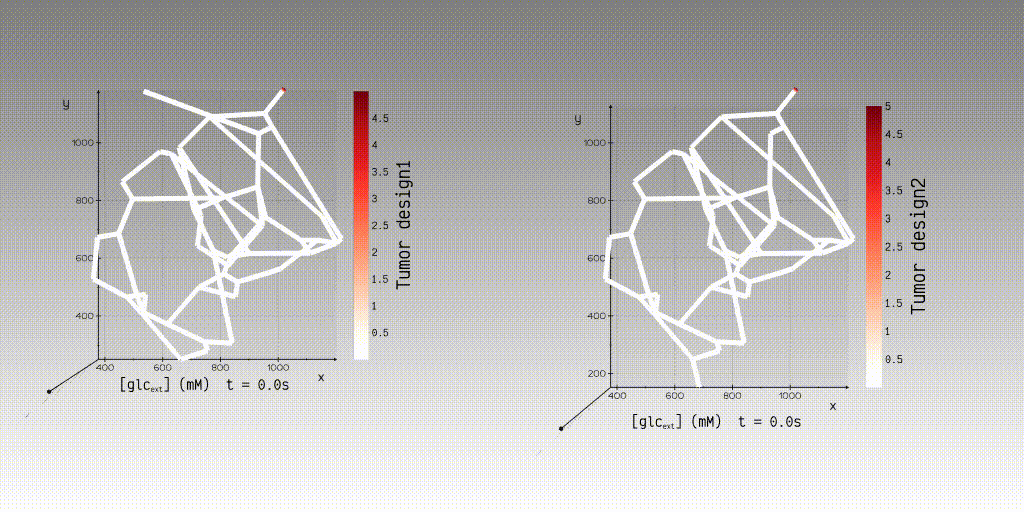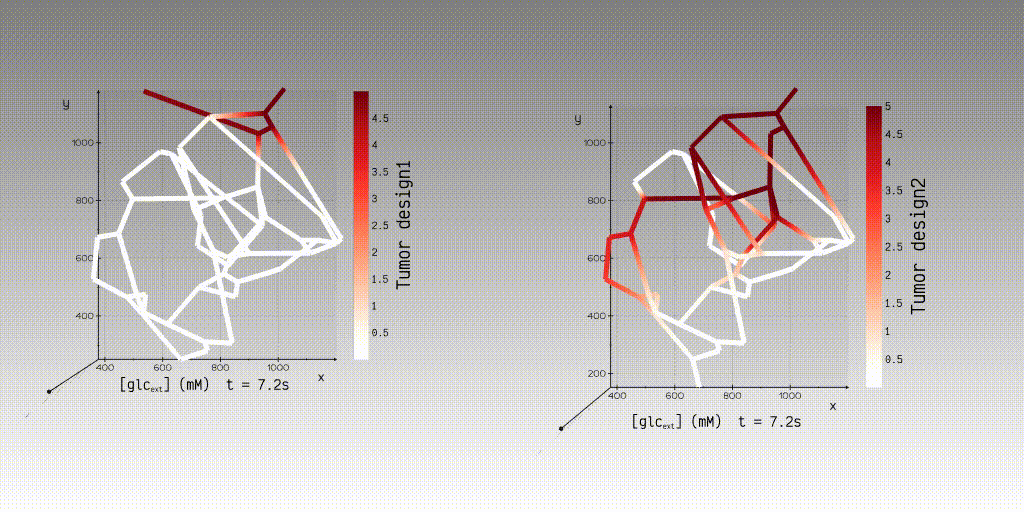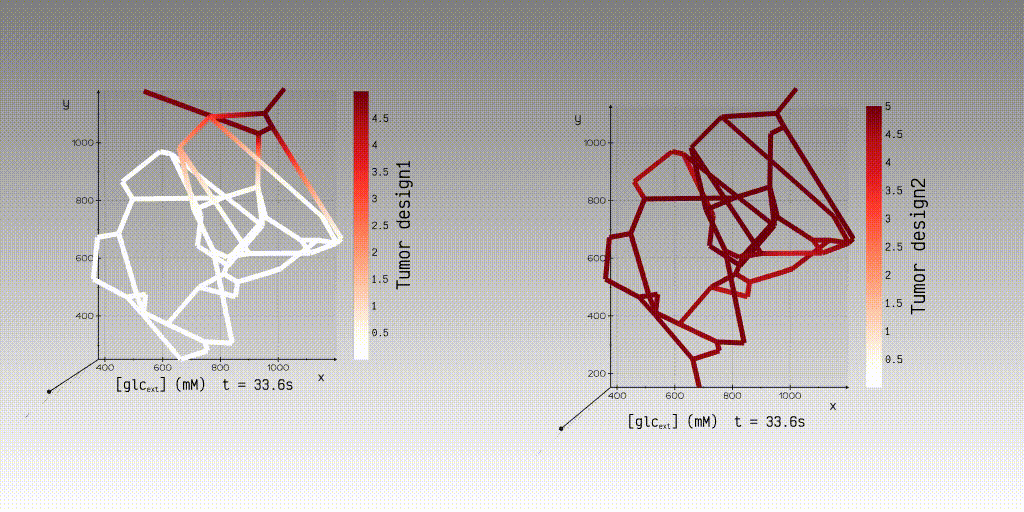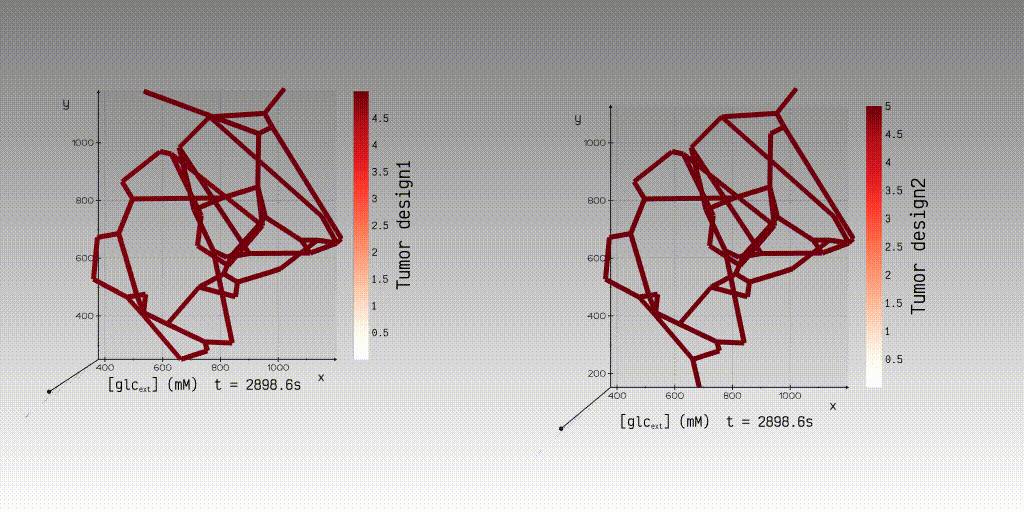
can use our application to simulate and view the simulation results to interactively examine the results of steady-state
velocity and pressure fields and dynamic concentration fields.