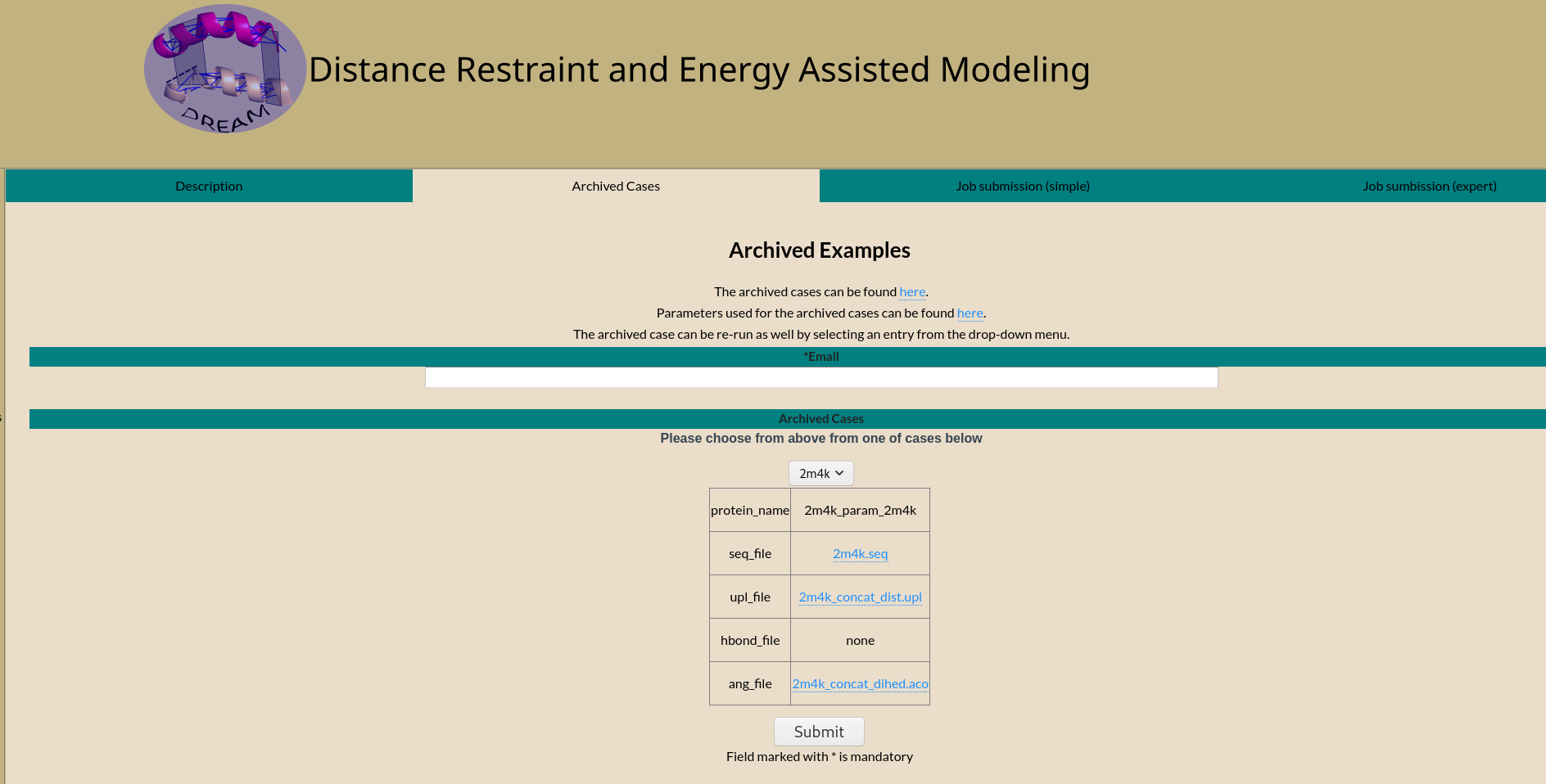
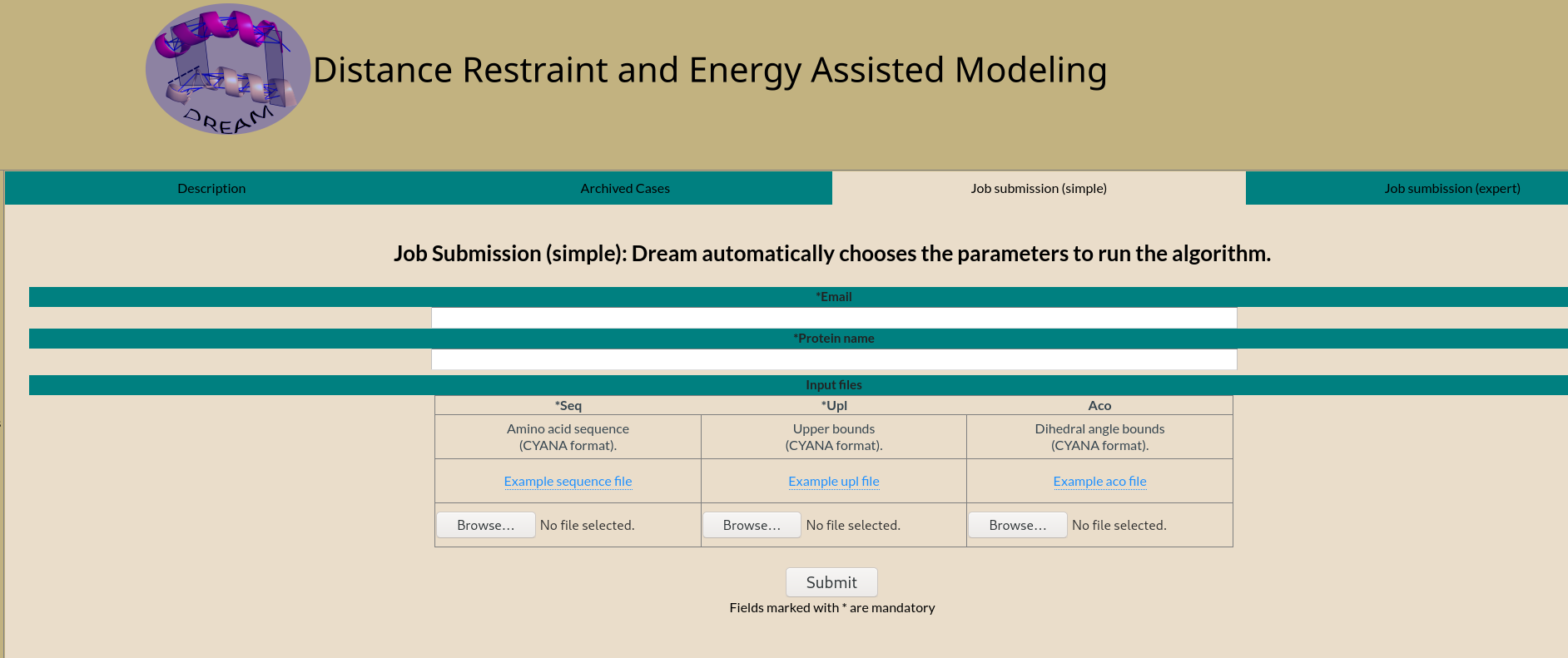
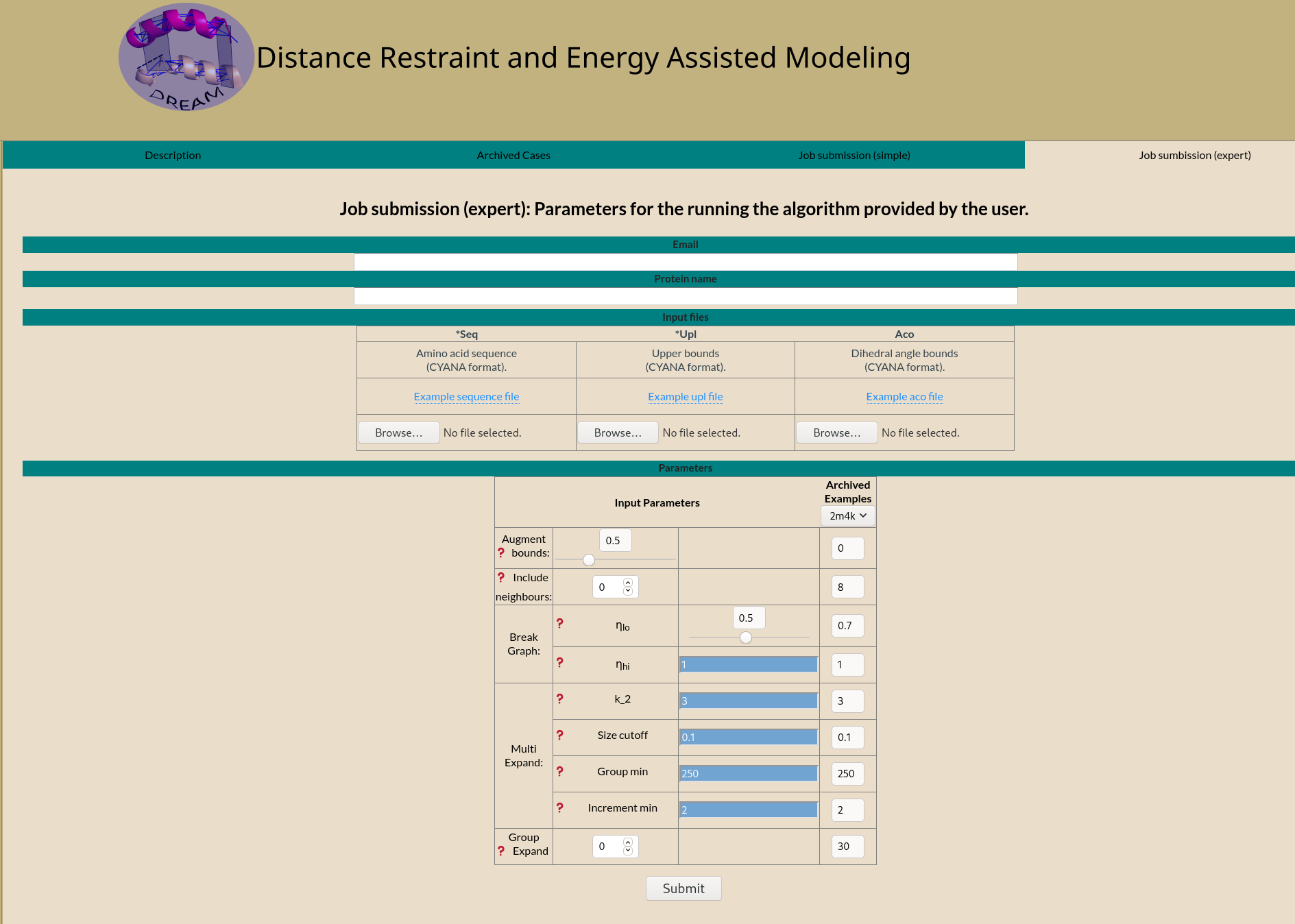
Emailed result
On completion an email is sent to the email address provided.
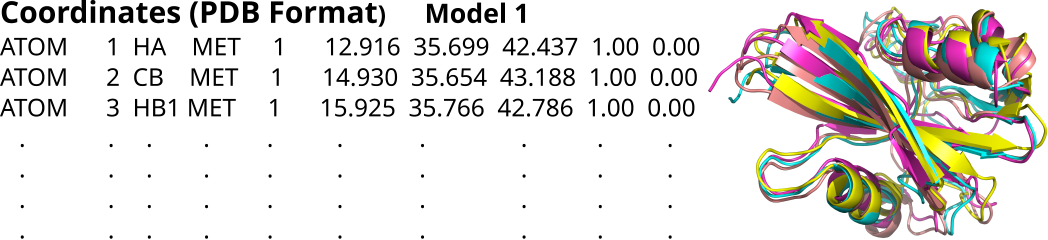
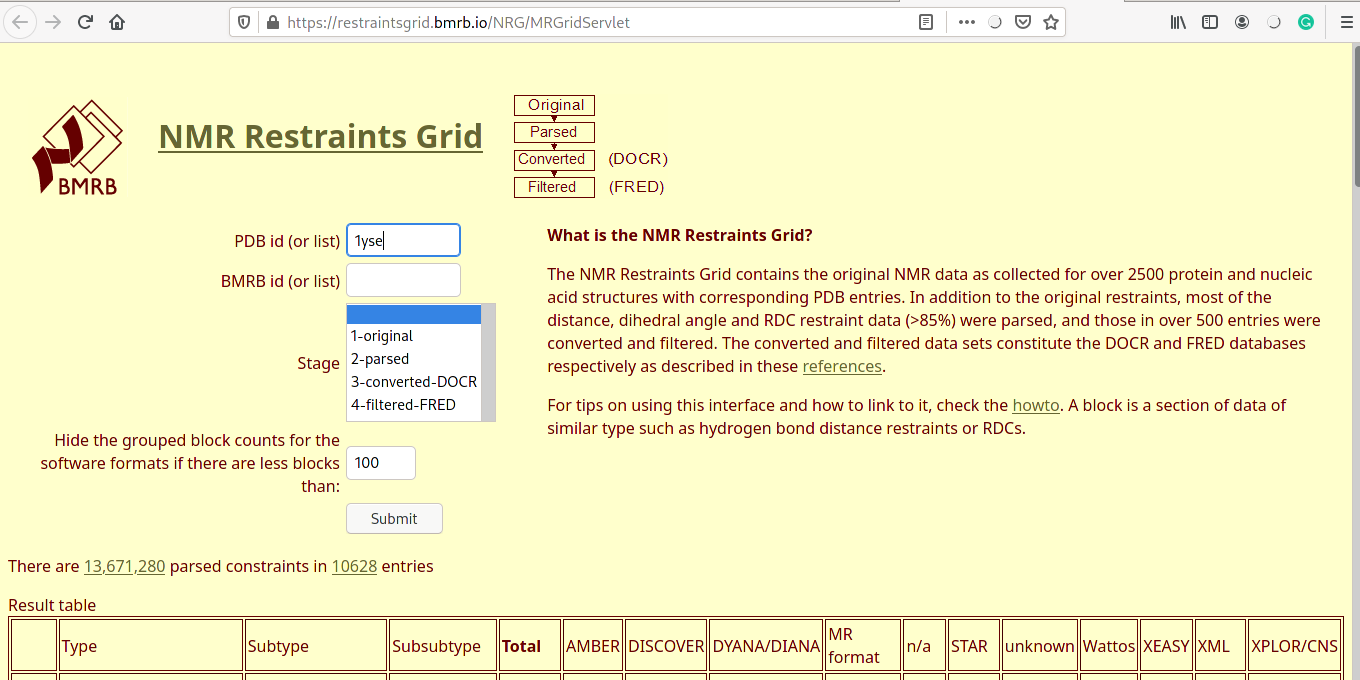
Protein structure files (PDB format)
- Individual files in PDB format for all the five models.
- Ensemble file containing all the models aligned together.
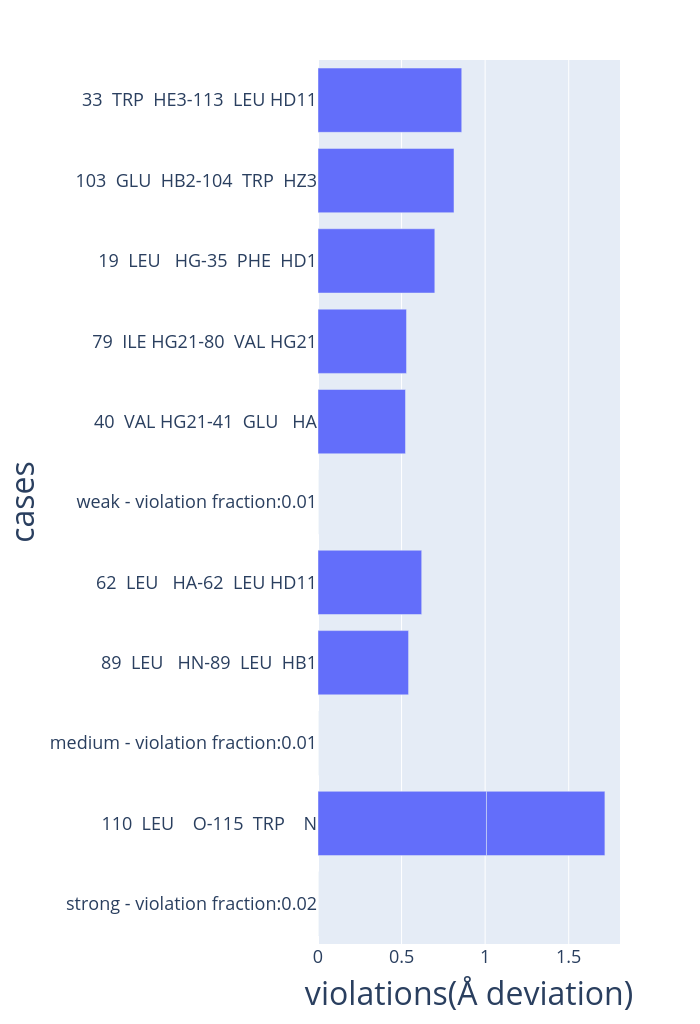
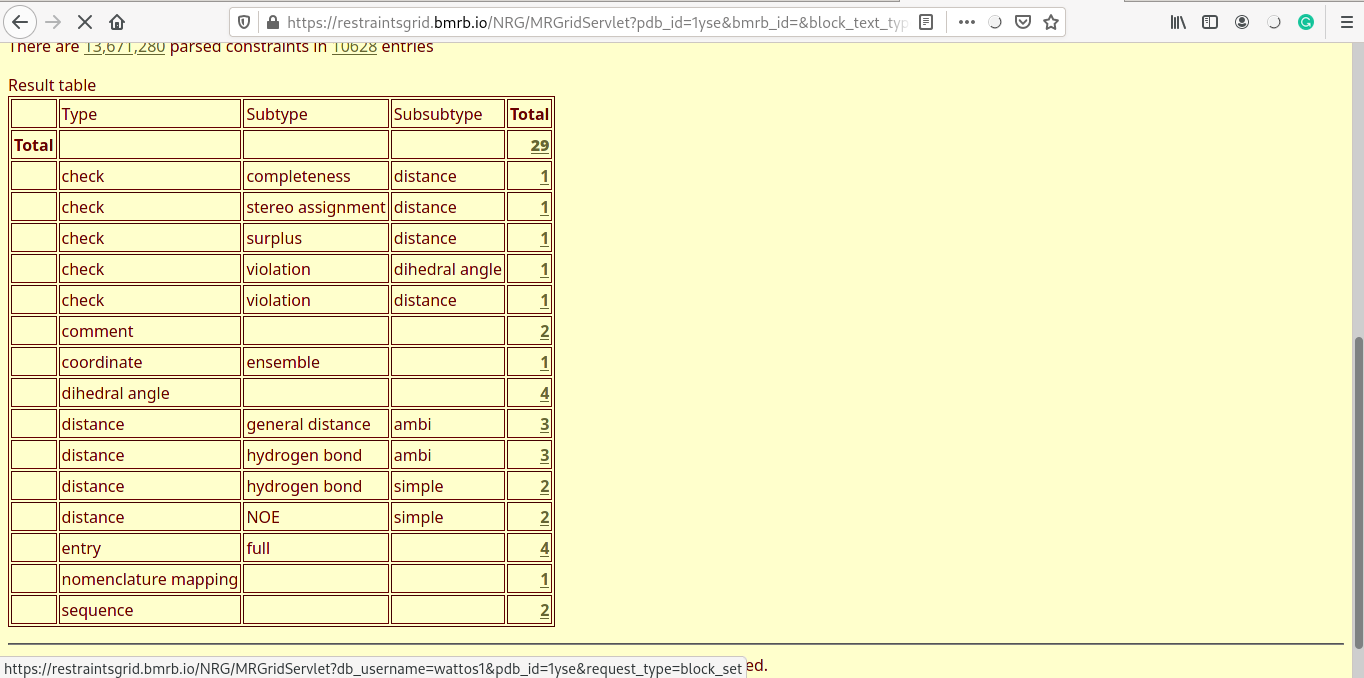
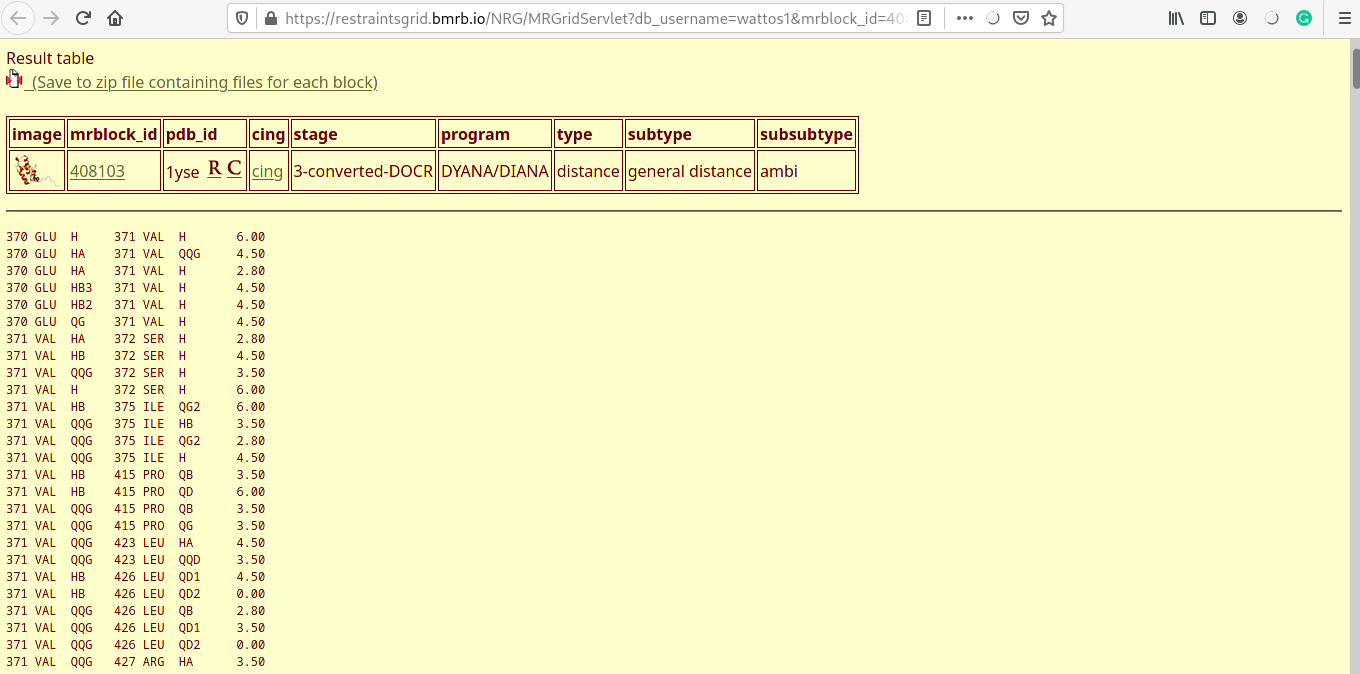
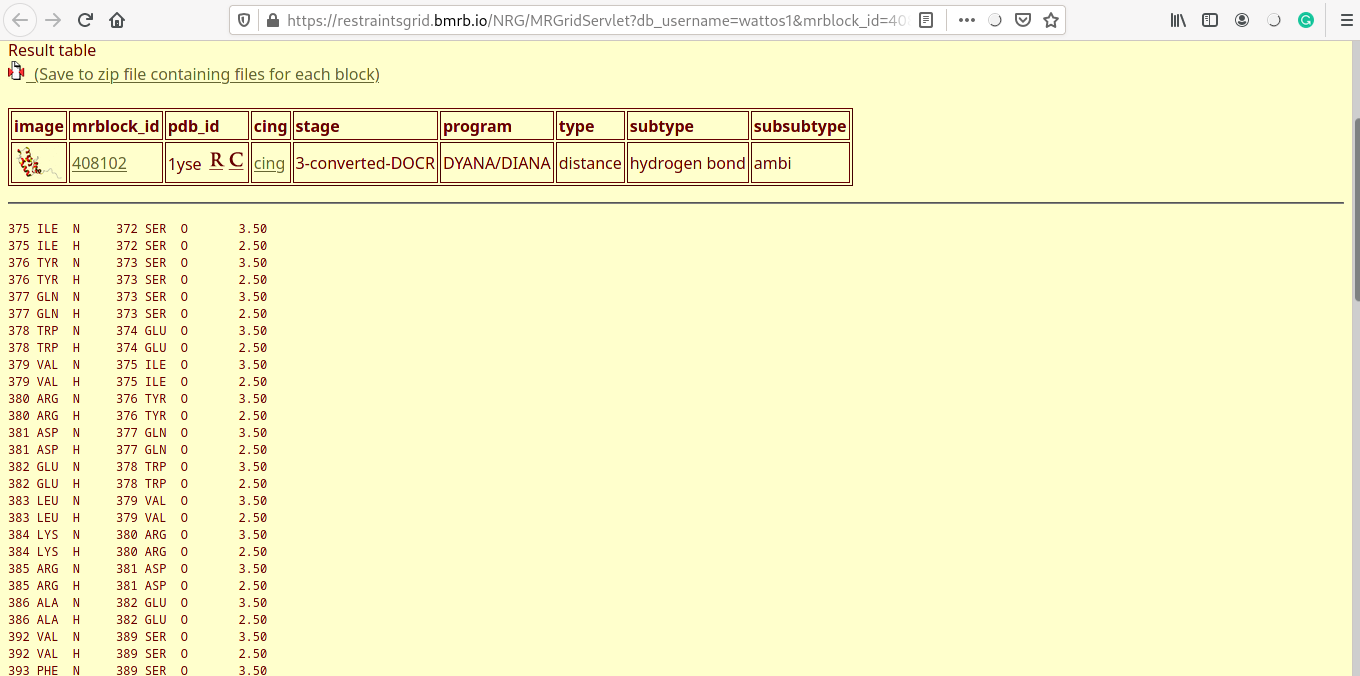
Experimental distance violations
- Distance violations for each of the five models are displayed (one shown here).
- Experimental distance costraints are categorised into strong (0-2.7Å), medium (2.7-3.5Å) and weak (3.5-6Å).
- Distance violations are reported for each of the categories, arranged in ascending order.
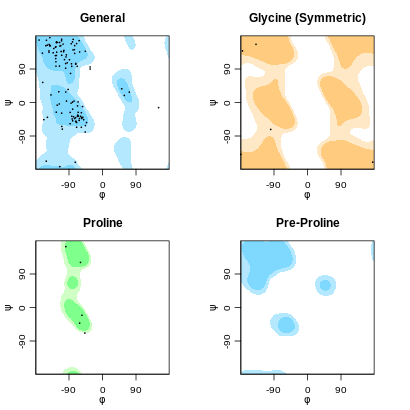
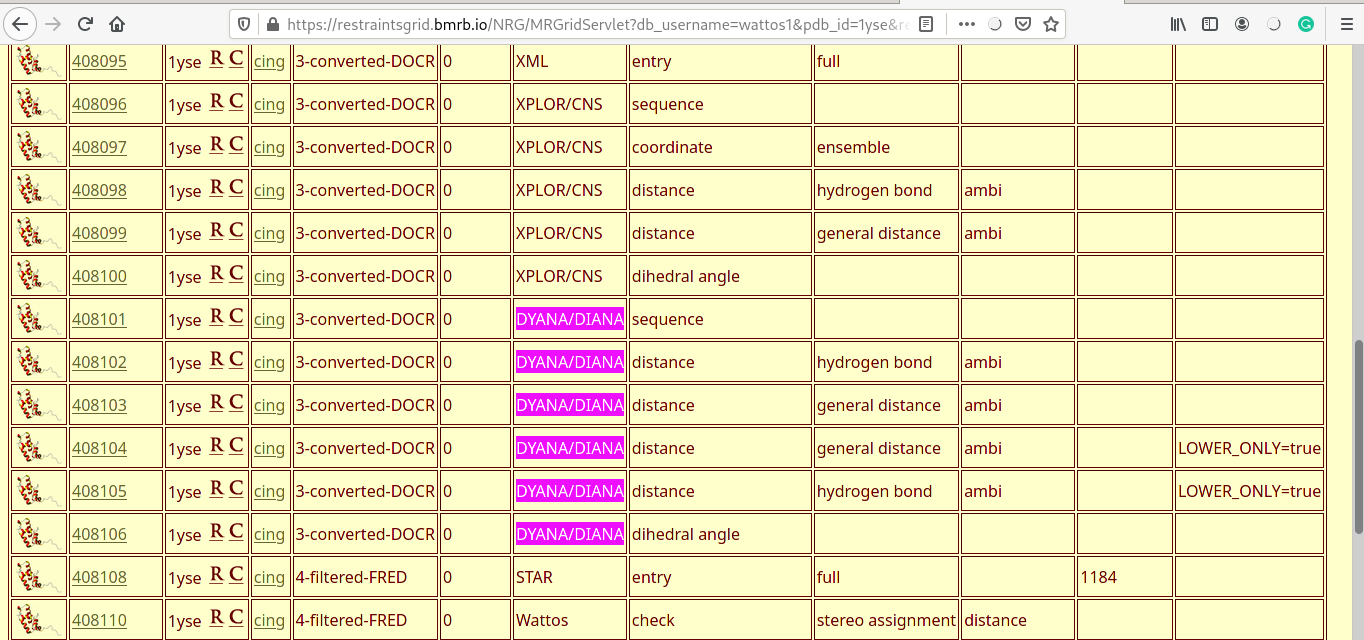
Ramachandran plot
- Distance violations for each of the five models are displayed (one shown here).
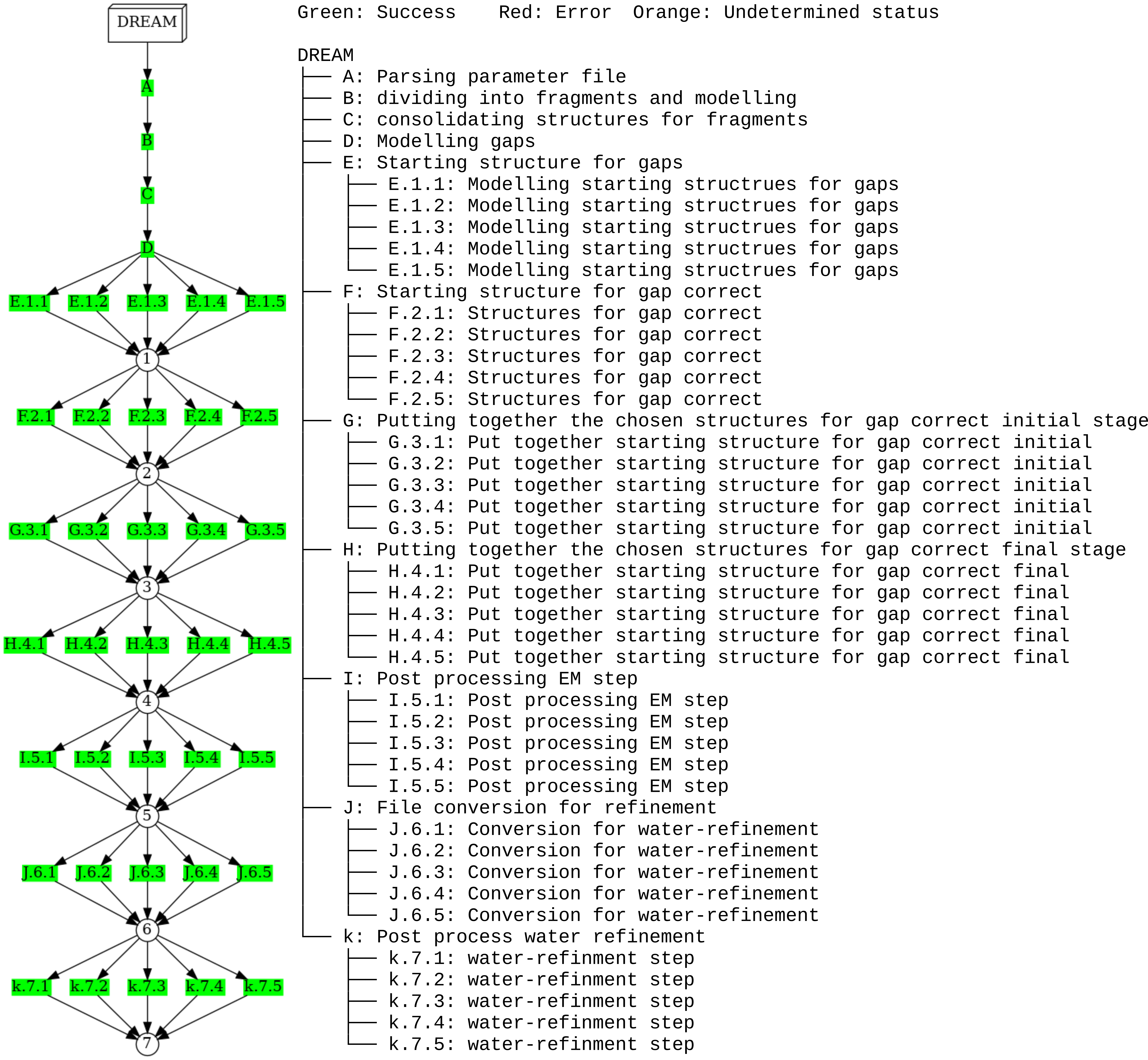
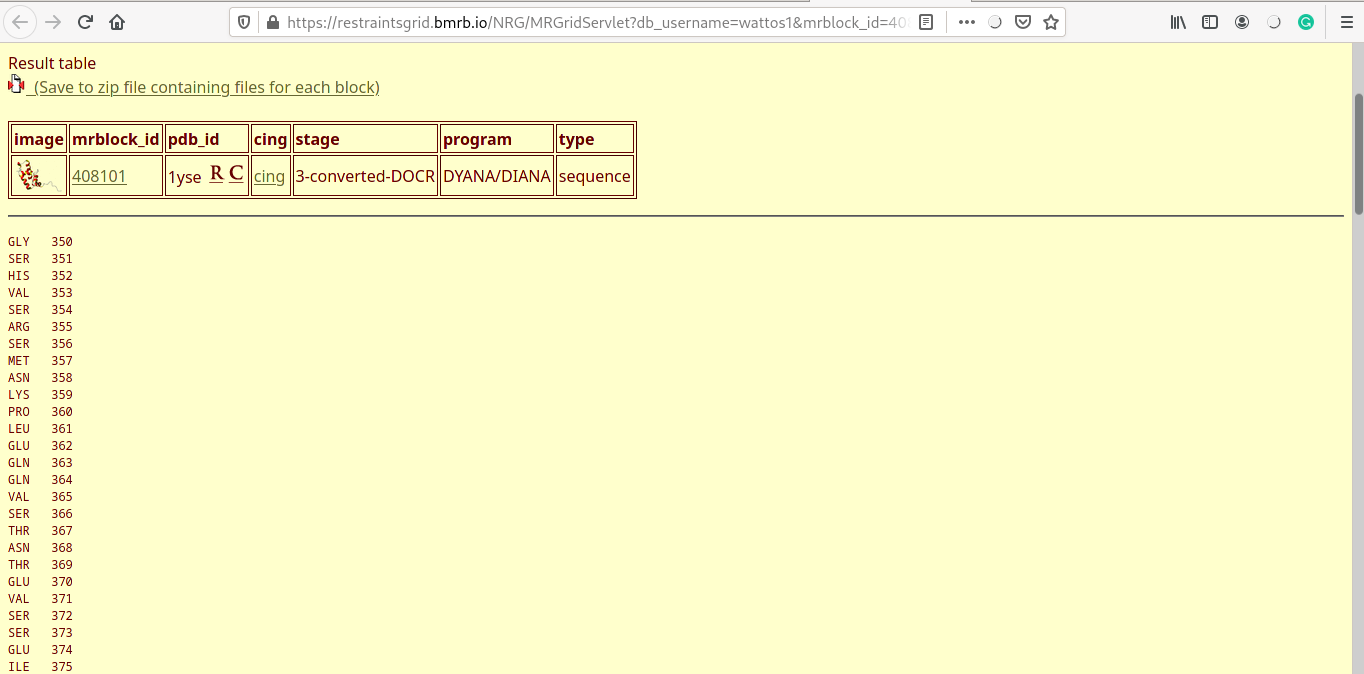
Progress for DREAM (archived examples and expert mode with parameters supplied)
- Progress of the DREAM algorithm, for running archived examples of expert mode with parameters supplied by the user.
- Steps such as "divide and model", "model gaps", "gap correct" and "post processing" steps proceeds in parallel.
- Various nodes in the graph are colour coded as green (success), orange (steps did not complete) and red (failoure).
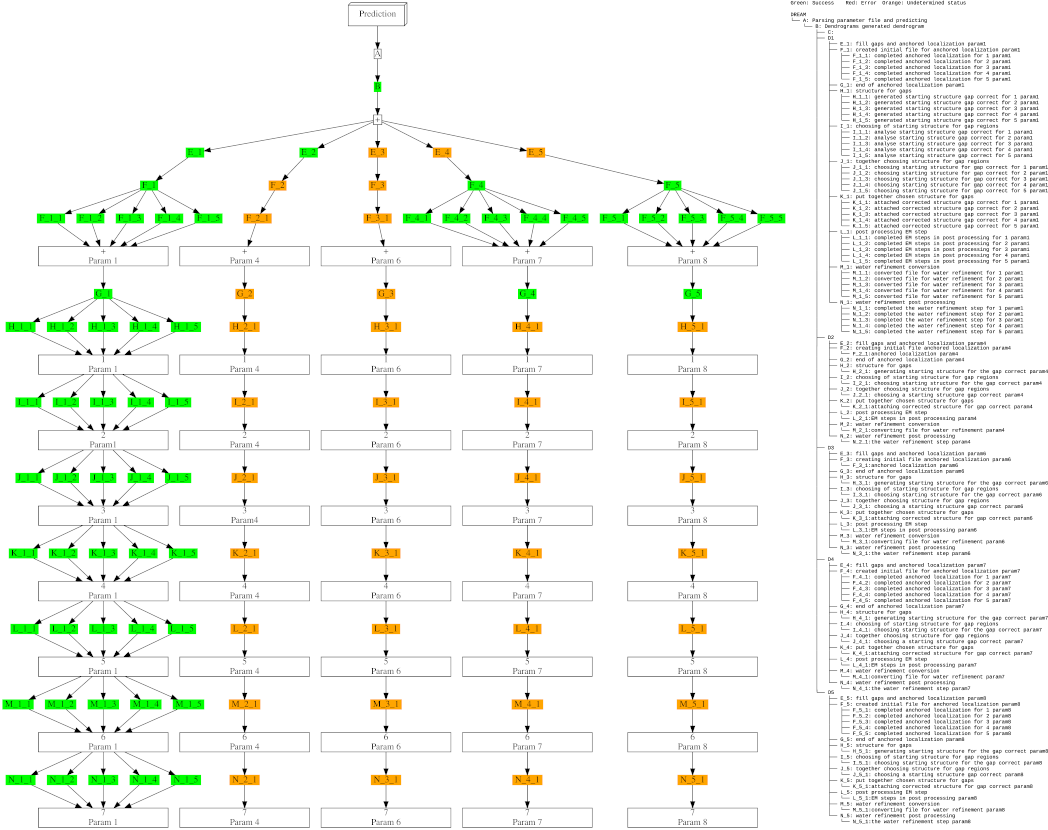
Progress for DREAM (simple-expert mode)
- Progress of the DREAM algorithm, for running in expert mode (simple).
- Four parameters were predicted to model the structure for the protein, namely "Param-1", "Param-4", "Param-6", "Param-7" and "Param-8"
- Using the parameter set in "Param-1", "Param-7" and "Param-8", were able to model structure for the fragment having divided the protein into regions having larger concentration of experimental bounds
- However, DREAM choose the parameter set, i.e., "Param-1" which gives largest coverage of the protein with fewer outliers in the ramachandran plot.
- Note that after parameter set "Param-1" is chosen, computation for other parameter set is stopped.
- Steps such as "divide and model", "model gaps", "gap correct" and "post processing" steps proceeds in parallel.
- Various nodes in the graph are colour coded as green (success), orange (steps did not complete) and red (failoure).
Back to top of page